Mapping the lipid-protein interface of the Torpedo californica and muscle-type acetylcholine receptors
Background
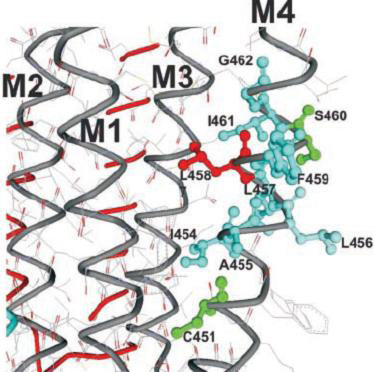
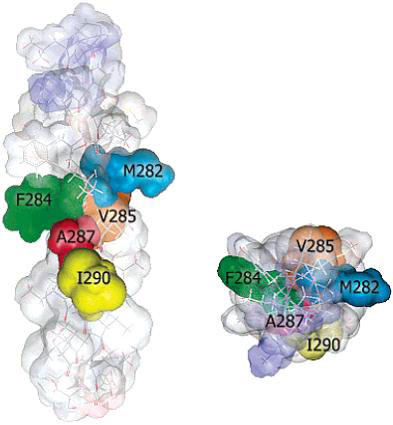
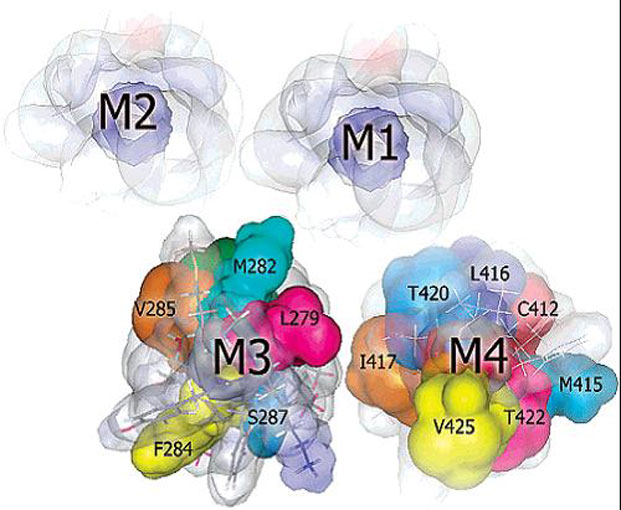
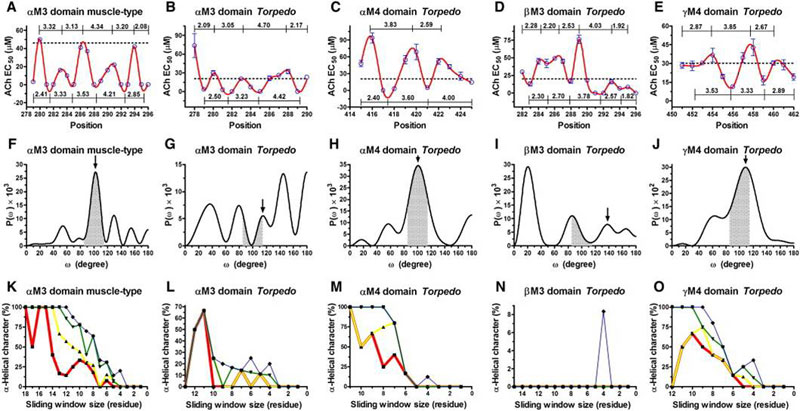
The nicotinic acetylcholine receptor (AChR) is a ligand-gated ion channel, integral membrane protein, which mediates fast chemical synaptic transmission at the neuromuscular junction and throughout the nervous system and is activated by binding of the chemical neurotransmitter acetylcholine (ACh). Topologically, AChR is a pentamerous receptor that consists of 20 putative transmembrane domains (4 transmembrane domains per AChR-subunit) of which 10 are lipid-exposed transmembrane domains (LETMDs) denoted as M3 and M4 domains. The studies that combined site-directed mutagenesis and electrophysiology (two-electrode voltage clamp and patch clamp) demonstrated that a single tryptophan substitution in the LETMDs could alter the kinetics rates, expression levels, and activatable fraction of the AChR. The extension of these studies along the LETMDs (tryptophan-scanning mutagenesis; TrpScanM) has revealed structure-function relationships, allosteric hydrophobic sites, helical structural models, transitional conformational changes, and functional and structural divergence between Torpedo and muscle-type AChRs and has provided the development of a hypothetical dynamic model (tilting-spring model). In addition, it develops a mathematical tool that combine the TrpScanM data with Fourier transform to reliably predict the helical structure of the LETMDs. In spite of valuable information about LETMDs provided by our laboratory, some LETMDs (ßM3, ßM4, ?M3, ?M4, dM3, dM4, eM3, and eM4 domains of the muscle-type AChR) have not been studied yet, keeping unrevealing structural and functional information of these LETMDs.
Long-term goal
Our long-term goal is to elucidate the role of lipid-protein interface of AChR in the cholinergic synapses of the nervous system.
Objective
The objective of this research project is to map the lipid-protein interface of the Torpedo and muscle-type AChRs. Hypothesis The central hypothesis of the research project is that the lipid-protein interface modulates AChR function and the fraction of receptors that could be activatable by ACh neurotransmitter.
Rationale
The rationale for the research project are to: (1) define the structure and spatial orientation of the LETMDs, (2) gain insight into the molecular basis for the lipid interaction and regulation of AChR function, (3) define the mechanistic link between LETMDs and gating machinery of the AChR, and (4) understand the structural basis for differential sensitivities of different AChRs species to the lipid environment.
Working objectives
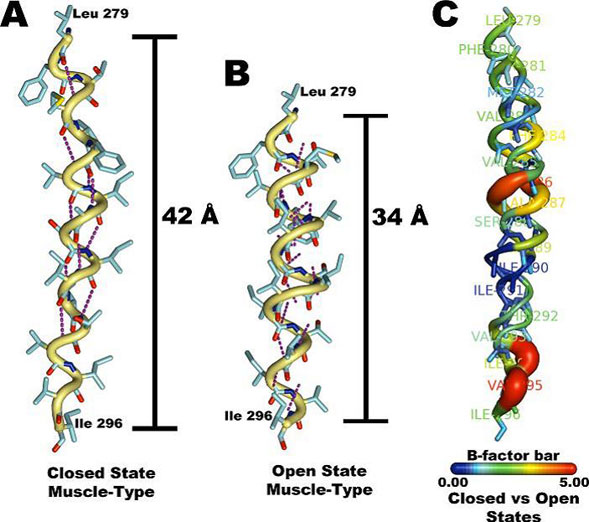
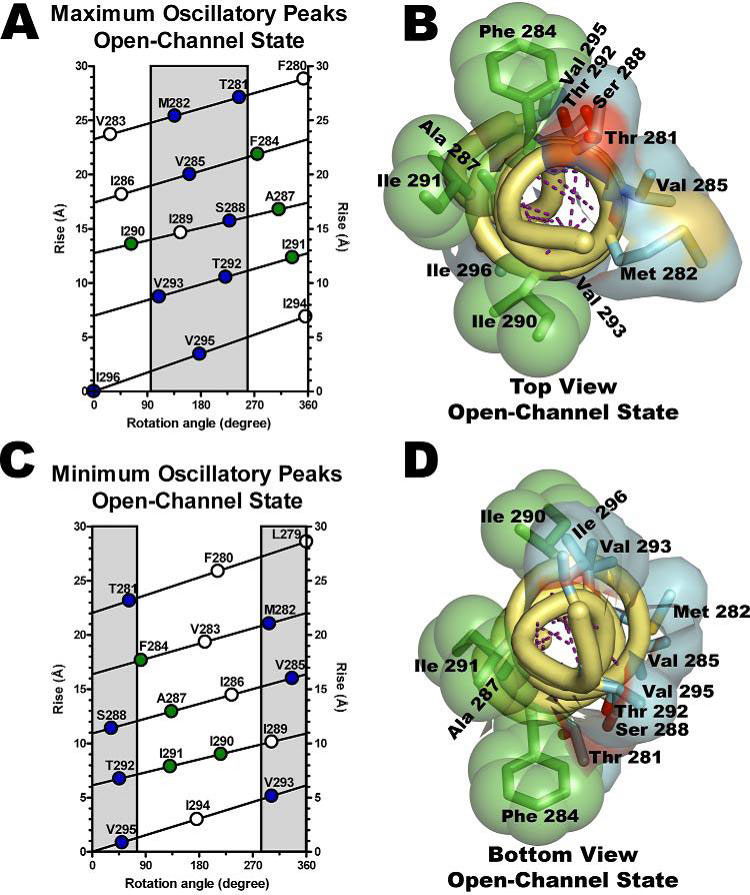
The objective of this project is to complete the TrpScanM data for all the M3 and M4 lipid-exposed domains of the muscle-type AChRs to develop structural models that will be compared with the Torpedo AChR and with the most recent AChR structure (Unwin et al., 2005). The structural models generated from TrpScanM data will be used to estimate: (1) spatial orientation of the helix relative to the interior of the protein, (2) overall secondary structure, (3) membrane crossing angle predictions, (4) altered patterns of hydrogen bonds inside the helix, (5) periodicity of the helix at the center and at the edge with the membrane, (6) potential deviations from helical structure, (7) prediction on the degree of lipid contact and (8) overall movement on each of the lipid exposed domains upon agonist activation. These parameters will be estimated for the closed and open states of the Torpedo and muscle-type AChRs. These structural models will be use to generate a detailed structural model of the AChR lipid interface for Torpedo and muscle-type AChRs.
References
Caballero-Rivera D, Cruz-Nieves OA, Oyola-Cintrón J, del Hoyo-Rivera N, Otero-Cruz JD, Torres-Nuñez DA & Lasalde-Dominicci JA (2008) Role of the lipid-exposed dM4 transmembrane domain on the function and structure of the Torpedo californica nicotinic acetylcholine receptor. (Manuscript in preparation).
Díaz-De León R, Otero-Cruz JD, Torres-Nuñez DA, Casiano A & Lasalde-Dominicci JA (2008) Tryptophan scanning of the acetylcholine receptor’s bM4 transmembrane domain; decoding allosteric linkage at the lipid-protein interface with ion-channel gating. Channels (Manuscript in revision).
Otero-Cruz JD, Torres-Núñez DA, Báez-Pagán CA & Lasalde-Dominicci JA (2008) Fourier transform coupled to tryptophan-scanning mutagenesis: Lessons from its application to the prediction of secondary structure in the acetylcholine receptor lipid-exposed transmembrane domains. BBA – Proteins & Proteomics 1784(9):1200-7.
Otero-Cruz JD, Báez-Pagán CA, Caraballo-González IM & Lasalde-Dominicci JA (2007) Tryptophan-scanning mutagenesis in the aM3 transmembrane domain of the muscle-type acetylcholine receptor. A spring model revealed. J Biol Chem 282(12):9162-71.
Guzmán GR, Ortiz-Acevedo A, Ricardo A, Rojas LV & Lasalde-Dominicci JA (2006) The polarity of lipid-exposed residues contributes to the functional differences between Torpedo and muscle-type nicotinic receptors. J Membr Biol 214(3):131-8.
Ortiz-Acevedo A, Melendez M, Asseo AM, Biaggi N, Rojas LV & Lasalde-Dominicci JA (2004) Tryptophan scanning mutagenesis of the ?M4 transmembrane domain of the acetylcholine receptor from Torpedo californica. J Biol Chem 279(40):42250-7.
Santiago J, Guzmán GR, Torruellas K, Rojas LV & Lasalde-Dominicci JA (2004) Tryptophan scanning mutagenesis in the TM3 domain of the Torpedo californica acetylcholine receptor beta subunit reveals an a-helical structure. Biochemistry 43(31):10064-70.
Navedo M, Nieves M, Rojas L & Lasalde-Dominicci JA (2004) Tryptophan substitutions reveal the role of nicotinic acetylcholine receptor a-TM3 domain in channel gating: differences between Torpedo and muscle-type AChR. Biochemistry 43(1):78-84.
Guzmán GR, Santiago J, Ricardo A, Martí-Arbona R, Rojas LV & Lasalde-Dominicci JA (2003) Tryptophan scanning mutagenesis in the aM3 transmembrane domain of the Torpedo californica acetylcholine receptor: functional and structural implications. Biochemistry 42(42):12243-50.
Cruz-Martín A, Mercado JL, Rojas LV, McNamee MG & Lasalde-Dominicci JA (2001) Tryptophan substitutions at lipid-exposed positions of the ?M3 transmembrane domain increase the macroscopic ionic current response of the Torpedo californica nicotinic acetylcholine receptor. J Membr Biol 183(1):61-70.
Tamamizu S, Guzmán GR, Santiago J, Rojas LV, McNamee MG & Lasalde-Dominicci JA (2000) Functional effects of periodic tryptophan substitutions in the aM4 transmembrane domain of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 39(16):4666-73.
Tamamizu S, Lee Y, Hung B, McNamee MG & Lasalde-Dominicci JA (1999) Alteration in ion channel function of mouse nicotinic acetylcholine receptor by mutations in the M4 transmembrane domain. J Membr Biol 170(2):157-64.
Ortiz-Miranda SI, Lasalde JA, Pappone PA & McNamee MG (1997) Mutations in the M4 domain of the Torpedo californica nicotinic acetylcholine receptor alter channel opening and closing. J Membr Biol 158(1):17-30.
Lasalde JA, Tamamizu S, Butler DH, Vibat CR, Hung B & McNamee MG (1996) Tryptophan substitutions at the lipid-exposed transmembrane segment M4 of Torpedo californica acetylcholine receptor govern channel gating. Biochemistry 35(45):14139-48.
Lee YH, Li L, Lasalde J, Rojas L, McNamee M, Ortiz-Miranda SI & Pappone P (1994) Mutations in the M4 domain of Torpedo californica acetylcholine receptor dramatically alter ion channel function. Biophys J 66(3 Pt 1):646-53.